Operational Synthesis
Learning Objectives
After completing this module, you will be able to:
- Summarize the advantages of creating a defined contribution workflow
- Explain how synthesis teams can use GitHub to collaborate more efficiently and reproducibly
- Identify characteristics of reproducible coding / project organization
- Explain benefits of reproducibility (to your team and beyond)
- Understand best practices for preparing and analyzing data to be used in synthesis projects
Introduction
Scientific research can be defined as “creative and systematic work undertaken in order to increase the stock of knowledge” (2015 Frascati Manual). To this basic definition of research, our definition of synthesis research adds collaborative work, and the integration and analysis of a wide range of data sources.
In Module 1 we discussed many of the collaborative considerations for synthesis research, including creating a diverse and inclusive team, asking synthesis-ready scientific questions (often broad in scope or spatial scale), and finding suitable information (or data) from a wide variety of sources to answer those questions.
Once the synthesis team moves into the operational phase of research, which includes the integration and analysis of data, there are some key activities that must happen:
- Make a plan for technical challenges
- Harmonize and tidy data in a reproducible way
- Analyze data to answer your questions
Once you have analyzed your data, you can interpret the results and create products as you likely would in a non-synthesis project–though the scale and impact of your products are likely to be larger for a synthesis project than for a typical scientific effort. More on these later steps of synthesis work in Module 3.
We’ve already seen that creating a collaborative, inclusive team can set the stage for successful synthesis research. Each of the operational activities above will also benefit from this mindset, and in this module we highlight some of the most important considerations and practices for a team science approach to the nuts-and-bolts of synthesis research.
1. Make a Plan
For non-synthesis projects, it can feel intuitive to skip this step. Especially if you are working alone or with a small group of close collaborators the formal process of making and documenting your strategy can feel like a waste of time relative to ‘actually’ doing the work. However, synthesis work uses a lot of data, requires a high degree of technical collaboration, and involves a series of big judgement calls that will need to be revisited. Because of these factors, taking the time to create an actual, specific, written plan (and periodically updating that plan as priorities and questions evolve) is vital to the success of synthesis projects!
See the sub-sections below for some especially critical elements to consider as you draft your plan. Note this is not exhaustive and there will definitely be other considerations that will be reasonable for your team to add to the plan based on your project’s goals and team composition.
How Will You Get (and Stay) Organized?
Two critical elements of oragnization are (1) the folder structure on your computer of project files and (2) how you will informatively name files so their purpose is immediately apparent. Also, it is easier to start organized than it is to get organized, so implementing a system at the start of your project and sticking to it will be much easier than trying to re-organize months or years worth of work after the fact.
While there is no “one size fits all” solution to project organization, we recommend something like the following:
Key properties of this structure:
- Everything nested within a project-wide folder
- Limited use of sub-folders
- Dedicated “README” files containing high-level information about each folder
- Consistent folder/file naming conventions
- Good names should be be both human and machine-readable (and sorted in the same way by machines and people)
- Avoid spaces and special characters
- Consistent use of delimeters (e.g., “-”, “_“, etc.)
- Shared file prefix (a.k.a. “slug”) connecting code files with files they create
- Allows for easy tracing of errors because the file with issues has an explicit tie to the script that likely introduced that error
- Includes a “data log” (a.k.a. “data inventory”) for documenting data sources and critical information about each
- E.g., dataset identifiers/DOIs, search terms used in data repository, provenance, spatiotemporal extent and granularity, etc.)
Suggested Project Structure:
synthesis_project
|– README.md
|– code
| |– README.md
| |– 01_harmonize.R
| L 02_quality-control.R
|– data
| |– README.md
| |– data-log.csv
| |– raw
| L tidy
| |– 01_data-harmonized.csv
| L 02_data-wrangled.csv
|– notes
|– presentations
L publications
|– README.md
|– community-composition
L biogeochemistry
How Will You Collaborate?
Once you’ve decided on your organization method, you’ll need to decide as a team how you will work together. It is critical that your whole team agrees to whatever method you come up with because it will result in a lot of unnecessary work if a subset of people do not follow the plan–and thus introduce disorganization and inconsistency that someone will have to spend time fixing later.
A good rule of thumb is that you should plan for “future you.” What collaboration methods will you in 6+ months thank ‘past you’ for implementing? It may also be helpful to consider the negative side of that question: what shortcuts could you take now that ‘future you’ will be unhappy with?
A huge part of deciding how you will collaborate is deciding how you will communicate as a team. When you work asynchronously, how will you tell others that you are working on a particular code or document file? This does not have to be high tech–an email or Slack message can suffice–but if you don’t communicate about the minutiae, you risk duplicating effort or putting in conflicting work.
Where Will Code Live?
Unlike the other planning elements, we (the instructors) feel there is a single correct answer to this: your code should live in a version control system. Broadly, “version control” systems track iterative changes to files. In addition to the specific, line-by-line changes to project files, contributions of each team member are tracked, and there are straightforward systems for ‘rolling back’ files to an earlier state if needed.

As the comic to the right shows–and as all scientists know from experience–you will have several (likely many) drafts of a given product before the finalized version. With a version control system, all the revisions in each draft are saved without needing to manually ‘re-save’ the file with a date stamp or version number in the filename. Version control systems provide a framework for preserving these changes without cluttering your computer with all of the files that precede the final version.
While there are several version control options, we recommend Git as the version control software and GitHub as the online platform for storing Git repositories in a shareable way. Note that there are viable alternatives to GitHub (e.g., GitLab, GitKraken, etc.) but (A) all of these rely on Git ‘under the hood’ and (B) in order to give an effective tutorial of version control in this course, we need to narrow our focus to just one of these website options. From here onwards, we’ll use focus on GitHub but it may be worthwhile for your team to investigate some of the GitHub alternatives as they will all offer similar functionality.
GitHub Tutorial
Given the time restrictions for this short course, we’ll only cover how you can engage with GitHub directly through its website today. However, this focus is actually a benefit to synthesis teams because learning how to engage with a Git repository via GitHub can enable collaborators who are less comfortable writing code to still fully contribute and stay abreast of the team’s progress.
There are a lot of Git/GitHub tutorials that already exist so, rather than add yet another variant to that list, we’ll instead work through part of the workshop created by the Scientific Computing team of the Long Term Ecological Research (LTER) Network Office. Note too that those materials include a detailed, step-by-step guide for working with Git/GitHub through RStudio, as well as some discussion of the project management tools that GitHub supports. As noted earlier, we won’t cover those parts of that workshop today, but they might be useful for you to revisit later.
The workshop materials we will be working through live here but for convenience we have also embedded the workshop directly into this short course’s website (see below).
2. Prepare Data
Synthesis efforts are intensely data-heavy by their nature; as such, you’ll need to spend more time and effort in the data preparation phase than you would in a non-synthesis project. This means that doing reproducible work pays huge dividends! Making one’s work “reproducible”–particularly in code contexts–has become increasingly popular but is not always clearly defined. For the purposes of this short course, we believe that reproducible work has the following properties:
- Contains sufficient documentation for those outside of the project team to navigate and understand the project’s contents
- Allows anyone to recreate the entire workflow from start to finish
- Leads to modular, extensible research projects; adding data from a new site, or a new analysis, should be relatively easy in a reproducible workflow
- Contains detailed metadata for all data products and prerequisite software
Metadata is “data about the data,” or information that describes who collected the data, what was observed or measured, when the data were collected, where the data were collected, how the observations or measurements were made, and why they were collected. Metadata provide important contextual information about the origin of the data and how they can be analyzed or used. They are most useful when attached or linked to the data being described, and data and related metadata together are commonly referred to as a dataset.
Metadata for ecological research data are well described in Michener et al (1997),1 but there are many other kinds of metadata with different purposes.2 If you are publishing a research dataset and have questions about metadata, ask a data manager for your project, or staff at the repository you are working with, for help. Either can typically provide guidance on creating metadata that will describe your data and be useful to the community (here is one example). We’ll return to the subject of metadata in Module 3.
Reproducible Coding
While the reproducibility guidelines identified above apply to many different facets of a project, there are a few more when considering reproducible code specifically. For example:
- All interactions with data should be done with code
- A version control system should be used to track changes to code
- All code should include non-coding “comments” to provide explanation/context
- Any necessary software libraries should be loaded explicitly at the start of each script
- Relative file paths that are agnostic to operating system
- E.g.,
file.path("data", "bees.csv")instead of"~/Users/me/Documents/synthesis_project/data/bees.csv"
- E.g.,
- Functions should be “namespaced” (if not already required by your coding language)
- E.g.,
dplyr::mutate()instead ofmutate()
- E.g.,
The above list is non-exhaustive and there are other conditions you may want to consider such as using custom functions for repeated operations or adding sequential numbers to script names that must be run in a particular order but generally following the above list will keep your code on the “more reproducible” side of things.
Reproducibility in Synthesis
Finally, there are a few additional considerations for reproducible synthesis work in particular. Judgement calls need to be made and agreed to as a team. However, group members should “defer to the doers.” If you have a very strong opinion that differs from the feeling of most of the rest of the group and will result in a lot of extra work, you should be ready to volunteer to do that work or allow those who will be responsible for the work to have a larger role in deciding the shape of that effort.
Reproducible synthesis work also requires dramatically more communication–especially around contribution guidelines and intellectual credit. It’s best to keep track of who contributed what, so that everyone gets credit. This can be challenging in practice and makes sense to start recording early and in a transparent way.
Data Preparation Overview
The scientific questions being asked in synthesis projects are usually broad in scope, and it is therefore common to bring together many datasets from different sources for analysis. The datasets selected for analysis (source data) may have been collected by different people, in different places, using different methods, as part of different projects, or all of the above. Typically you will need to harmonize these disparate datasets by standardizing data structure, units of measurement, and file format. Once this is done, you’ll move on to data cleaning where you can check for malformed values, unreasonable rows, and filter out unwanted observations.
These processes can be easy or difficult depending on the quality of the source data, the differences between source data, and how much metadata (see callout above) is available to understand them. Also, note that you could clean each input dataset separately first, then harmonize the data, but it may be easier to harmonize first. For example, if you have 10 datasets, each with some type of sampling date column, it will be much easier to filter out particular dates if all that information is in the same format in the same column as opposed to working with whatever idiosyncratic formats and/or column names are present in each original file.
Harmonize Data
Data harmonization is the process of bringing different datasets into a common format for analysis. The harmonized data format chosen for a synthesis project depends on the source data, analysis plans, and overall project goals. It is best to make a plan for harmonizing data before analysis begins, which means discussing this with the team in the early stages of a synthesis project. As a general rule, it is also wise to use a scripted workflow that is as reproducible as possible to accomplish the harmonization you need for your project. Following this guidance lets others understand, check, and adapt your work, and will also make it much, much easier to bring new data and analysis methods into the project.
Data harmonization is hard work that sometimes requires trial and error to arrive at a useful end product. At the end of this section are some additional data harmonization resources to help you get started. Looking at a simple example might also help.
A Word About Harmonized Data Formats
Above, we have discussed several aspects of selecting a data format. There are at least three related, but not exactly equivalent, concepts to consider when formatting data. First, formats describe the way data are structured, organized, and related within a data file. For example, in a tabular data file about biomass, the measured biomass values might appear in one column, or in muiltiple columns. Second, the values of any variable can be represented in more than one format. The same date, for example, could be formatted using text as “July 2, 1974” or “1974-07-02.” Third, format may refer to the file format used to hold data on a disk or other storage medium. File formats like comma separated value text files (CSV), Excel files (XLSX), JPEG images, are commonly used for research data, and each has particular strengths for certain kinds of data.
A few guidelines apply:
- For formatting a tabular dataset, err towards simpler data structures, which are usually easier to clean, filter, and analyze
- Keeping data in “long format”,3 is one common recommendation for this
- When choosing a file format, err towards open, non-proprietary file formats that more people know and have access to
- Delimited text files, such as CSV files, are a good choice for tabular data
- Use existing community standards for formatting variables and files as long as they suit your project methods and scientific goals
- Using ISO standards for date-time variables, or species identifiers from a taxonomic authority, are good examples of this practice
- There is no perfect data format!
- Harmonizing data always involves some judgement calls and tradeoffs
When choosing a destination format for the harmonized data for a synthesis project, the audience and future uses of the data are also an important consideration. Consider how your synthesis team will analyze the data, as well as how the world outside that team will use and interact with the data once it is published. Again, there is no one answer, but below are a few examples of harmonized destination formats to consider.
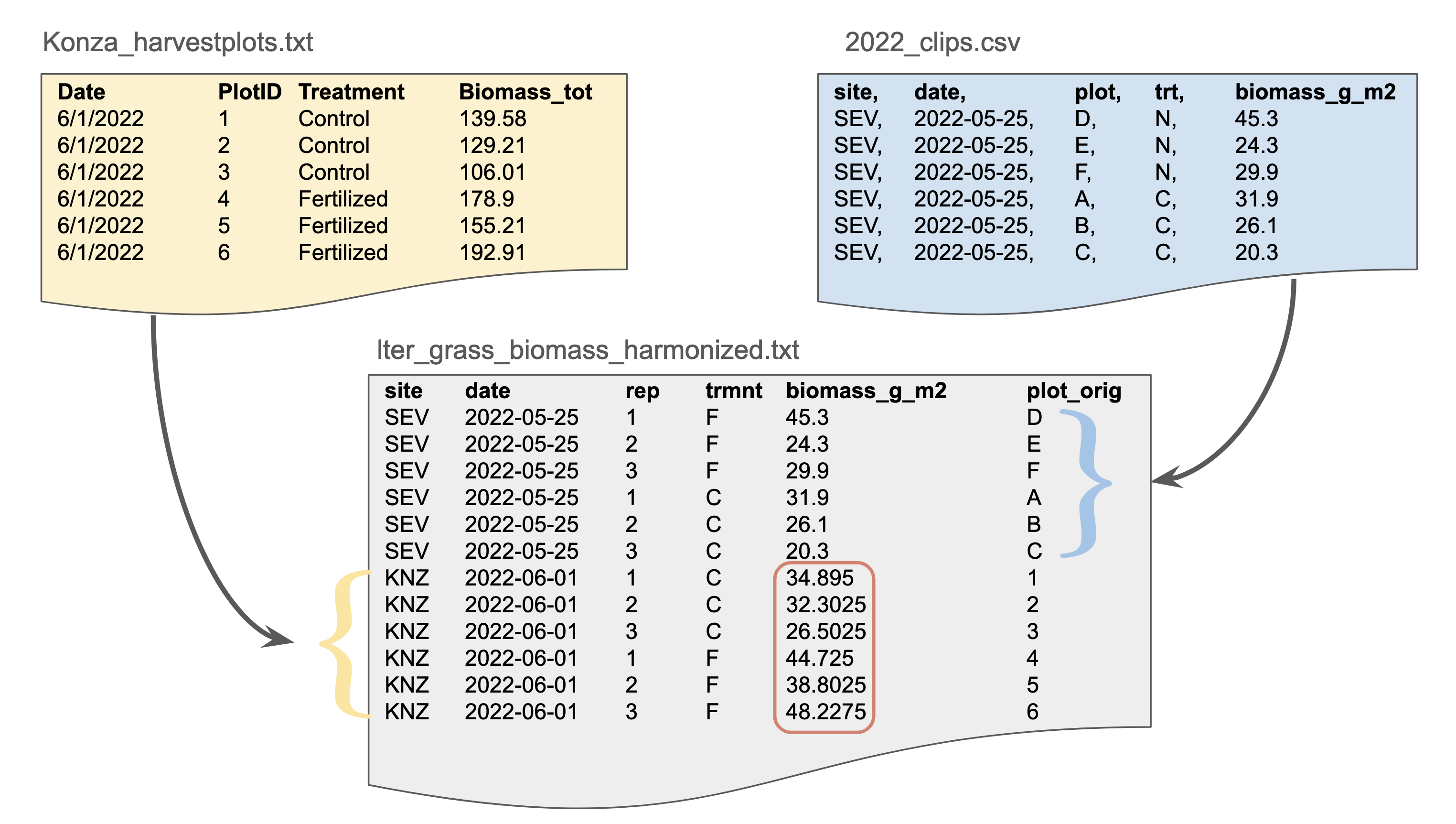
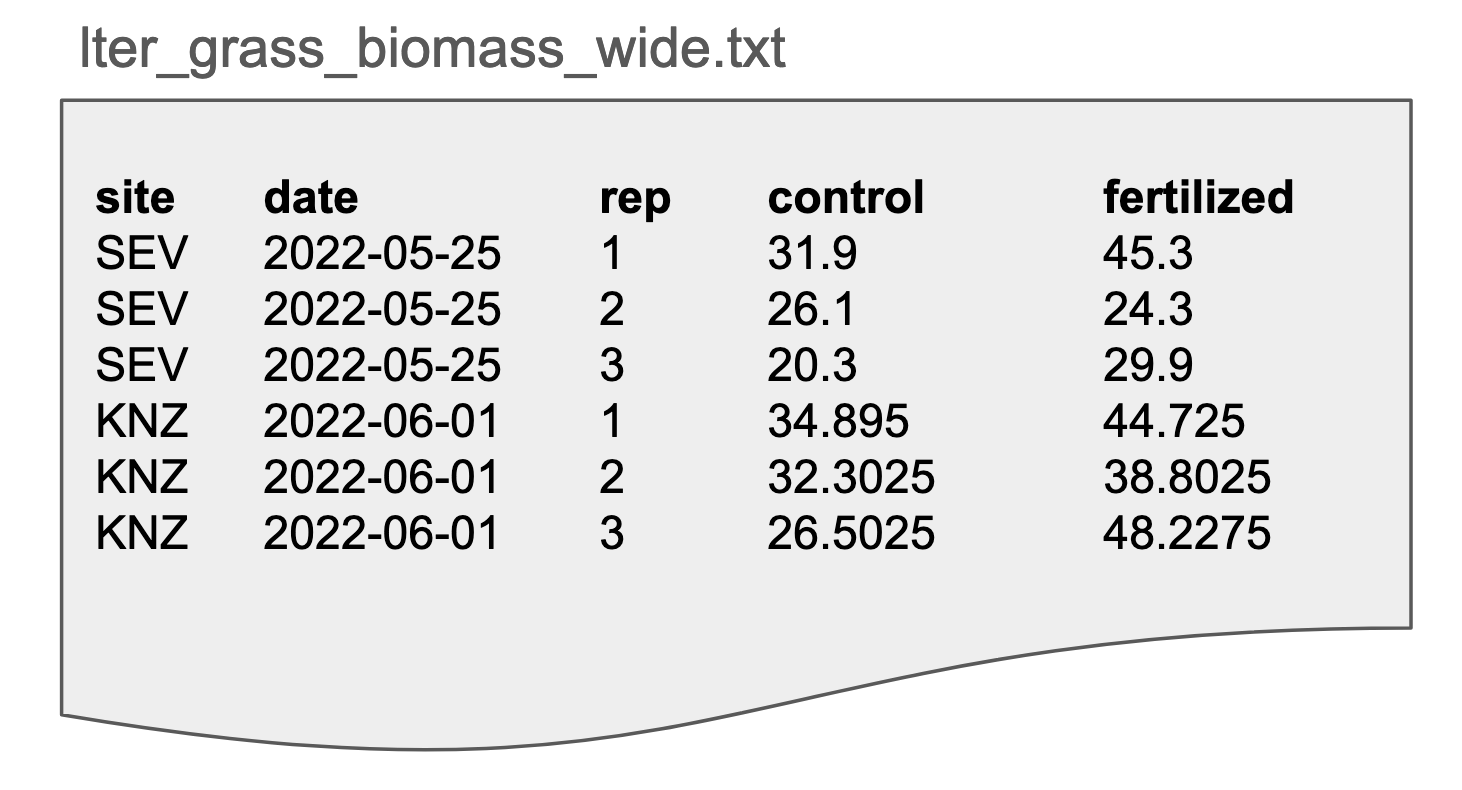
Here our grassland biomass data is in long format, often referred to as “tidy” data. Data in this format is generally easy to understand and use. There are three rules for tidy data:
- Each column is one variable.
- Each row is one observation.
- Each cell contains a single value.

Advantages: clear meaning of rows and columns; ease in filtering/cleaning/appending
Disadvantages: not as human-friendly so it can be difficult to assess the data visually
Possible file formats: Delimited text (tab delimited shown here), spreadsheets, database tables
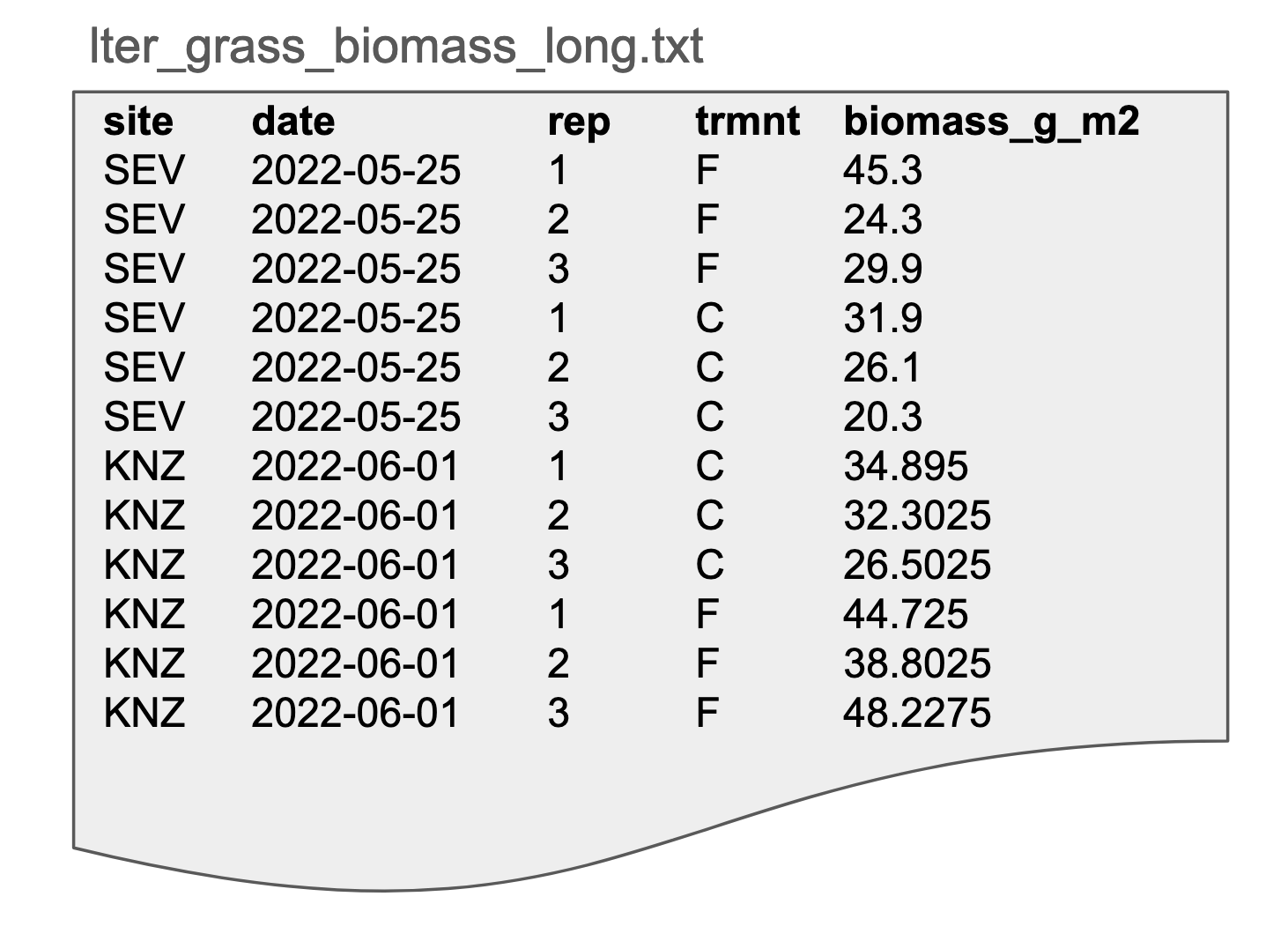
In this dataset, our grassland data has been restructured into wide format, often referred to (sometimes unfairly) as “messy” or “untidy” data. Note that the biomass variable has been split into two columns, one for control plots and one for fertilized plots.
Advantages: easier for some statistical analyses; easier to assess the data visually
Disadvantages: may be more difficult to clean/filter/append, multiple observations per row; more likely to contain empty (NULL) cells
Possible file formats: Delimited text (tab delimited shown here), spreadsheets, database tables
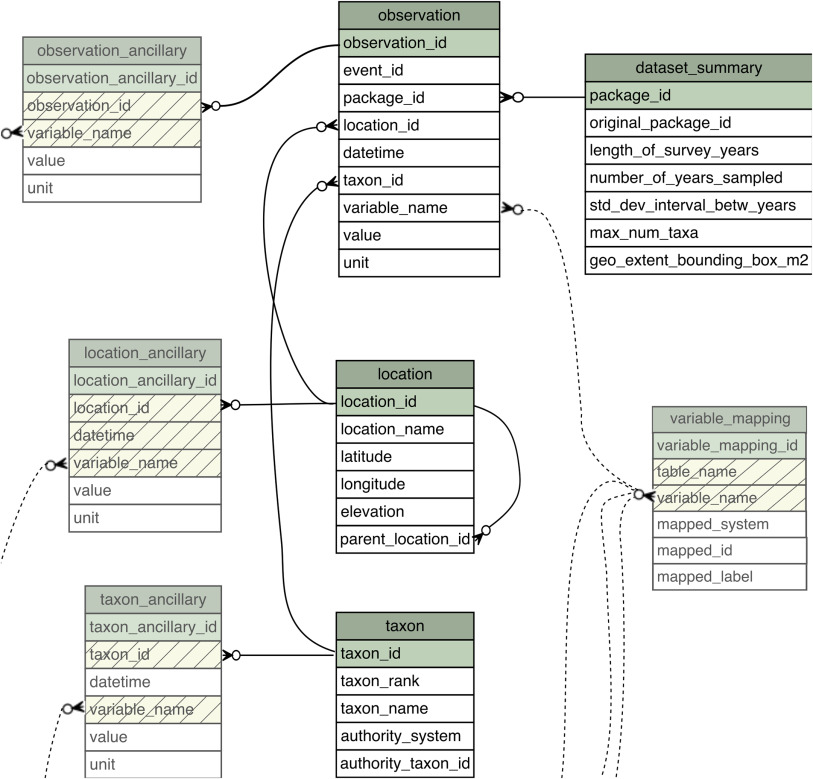
Below is an example of how we might structure our grassland data in a relational database. The schema consists of three tables that house information about sampling events (when, where data were collected), the plots from which the samples are collected, and the biomass values for each collection. The schema allows us to define the data types (e.g., text, integer), add constraints (e.g., values cannot be missing), and to describe relationships between tables (keys). Relational formats are normalized to reduce data redundancy and increase data integrity, which can help us to manage complex data4.

Advantages: reduced redundancy, greater integrity; community standard; powerful extensions (e.g., store and process spatial data); many different database flavors to meet specific needs
Disadvantages: significant metadata needed to describe and use; more complex to publish; learning curve
Possible file formats: Database stores, can be represented in delimited text (CSV)
A richer example is a schematic of the related tables that comprise the ecocomDP5 harmonized data format for biodiversity data. Eight tables are defined, along with a set of relationships between tables (keys), and constraints on the allowable values in each table.
There are many possibilities to make large synthesis datasets available and useful in the cloud. These require specialized knowledge and tooling, and reliable access to cloud platforms.
Advantages: easier access to big (high volume) data, can integrate with web apps
Disadvantages: less familiar/accessible to many scientists, few best practices to follow, costs can be higher
Possible file formats: Parquet files, object storage, distributed/cloud databases
There are many, many other possible harmonized data formats. Here are a few possible examples:
- DarwinCore archives for biodiversity data
- Organismal trait databases
- Archives of cropped, labeled images for training machine or deep learning models
- Libraries of standardized raster imagery in Google Earth Engine
Clean Data
When assembling large datasets from diverse sources, as in synthesis research, not all the source data will be useful. This may be because there are real or suspected errors, missing values, or simply because they are not needed to answer the scientific question being asked (wrong variable, different ecosystem, etc.). Data that are not useful are usually excluded from analysis or removed altogether. Data cleaning tends to be a stepwise, iterative process that follows a different path for every dataset and research project. There are some standard techniques and algorithms for cleaning and filtering data, but they are beyond the scope of this course. Below are a few guidelines to remember, and more in-depth resources for data cleaning are found at the end of this section.
- Always preserve the raw data. Chances are you’ll want to go back and check the original source data at least once.
- Use a scripted workflow to clean and filter the raw data, and follow the usual rules about reproducibility (comments, version control, functionalization).
- Consider using the concept of data processing “levels,” meaning that defined sets of data flagging, removal, or transformation operations are applied consistently to the data in stepwise fashion. For example, incoming raw data would be labeled “level 0” data, and “level 1” data is reached after the first set of processing steps is applied.
- Spread the data cleaning workload around! Data cleaning typically demands a huge fraction of the total time devoted to working with data,678 and it can be tedious work. Make sure the team shares this workload equitably.
A Word About Filtering Data
When filtering data, you are removing rows that you do not want to include in analysis. However, “filtering” is not a monolith and can be split into two categories that warrant different strategies. Some data are unusable and are removed for that reason, however, some data are potentially usable, but aren’t relevant to the current analysis.
Because of this latter category, filtering data should be done as late as possible in your workflow, ideally, as the final step before analysis. The reason for this is that you will eventually finish testing your first hypothesis and then the criteria for ‘is this data usable’ will change! If the very first step of your workflow as removing all data you didn’t want in that moment, you might have to return all the way to the start in order to get the data you now need for this subsequent hypothesis.
If instead, you develop your workflow with as much of the usable data as possible, when your filtering criteria evolve, most of your workflow–i.e., the labor-intensive harmonization and cleaning steps–is already good as-is and likely won’t need to be revisited. You simply create a new ‘filter’ script with the new criteria and can immediately leap into analysis.
3. Analyze Data
Once the team has found sufficient source data, then harmonized, cleaned, and filtered countless datasets, and documented and described everything with quality metadata, it is finally time to analyze the data! Great! Load up R or Python and get started, and then tell us how it goes. We simply don’t have enough time to cover all the ins and outs of data analysis in a this course. However, we have put a few helpful resources below to get you started, and many of the best practices we have talked about, or will talk about, apply:
- Document your analysis steps and comment your code, and generally try to make everything reproducible.
- Use version control as you analyze data.
- Give everyone a chance! Analyzing data is challenging, exciting, and a great learning opportunity. Having more eyes on the analysis process also helps catch interesting results or subtle errors.
Synthesis Group Case Studies
To make some of these concepts more tangible, let’s consider some case studies. The following tabs contain GitHub repositories for real teams that have engaged in synthesis research and chosen to preserve and maintain their scripts in GitHub. Each has different strengths and you may find that facets of each feel most appropriate for your group to adopt. There is no single “right” way of tackling this but hopefully parts of these exemplars inspire you.
LTER SPARC Group: Soil Phosphorus Control of Carbon and Nitrogen
Stored their code here: lter / lter-sparc-soil-p
Highlights
- Straightforward & transparent numbering of workflow scripts
- File names also reasonably informative even without numbering
- Simple README in each folder written in human-readable language
- Custom
.gitignoresafety net- Controls which files are “ignored” by Git (prevents accidentally sharing data/private information)
LTER Full Synthesis Working Group: The Flux Gradient Project
Stored their code here: lter / lterwg-flux-gradient
Highlights
- Extremely consistent file naming conventions
- Strong use of sub-folders for within-project organization
- Top-level README includes robust description of naming convention, folder structure, and order of scripts in workflow
- Active contribution to code base by nearly all group members
- Facilitated by strong internal documentation and consenus-building prior to choosing this structure
LTER Full Synthesis Working Group: From Poles to Tropics: A Multi-Biome Synthesis Investigating the Controls on River Si Exports
Stored their code here: lter / lterwg-silica-spatial
Highlights
- Files performing similar functions share a prefix in their file name
- Use of GitHub “Release” feature to get a persistent DOI for their codebase
- Separate repositories for each manuscript
- Nice use of README as pseudo-bookmarks for later reference to other repositories
For more information about LTER synthesis working groups and how you can get involved in one, click here.
Additional Resources
Courses, Workshops, and Tutorials
- Synthesis Skills for Early Career Researchers (SSECR) course. 2025. LTER Network Office
- Collaborative Coding with GitHub workshop. 2025. LTER Scientific Computing team
- Coding in the Tidyverse workshop. 2025. LTER Scientific Computing team
- R / Python Bilingualism tutorial. 2025. Nick J Lyon
- R Programming for Biologists (300-level undergraduate) course. 2024. Nick J Lyon
- Reproducible Approaches to Arctic Research Using R workshop. 2024. Arctic Data Center & NCEAS Learning Hub
- Shiny Apps for Sharing Science workshop. 2022. Lyon, N.J. et al.
- Ten Commandments for Good Data Management. 2016. McGill, B.
Literature
- Lyon, N.J. & Earl, S. Conventional Commits: A better way to track changes with Git. 2024. DataBits
- Todd-Brown, K.E.O., et al. Reviews and Syntheses: The Promise of Big Diverse Soil Data, Moving Current Practices Towards Future Potential. 2022. Biogeosciences
- Borer, E.T. et al. Some Simple Guidelines for Effective Data Management. 2009. Ecological Society of America Bulletin
Data Preparation
For R and Python users, there are excellent documentation resources that thoroughly cover data harmonization techniques like data filtering, reformatting, joins, and standardization.
- In Hadley Wickham’s R for Data Science book, the chapters on data transforms and data tidying are a good place to start
- Also for R users, the LTER Network’s
ltertoolspackage has a nice approach to harmonization with its intuitively-namedharmonizefunction. See theltertoolspackage vignette for details - In Wes McKinney’s Python for Data Analysis book, the chapter on data wrangling is helpful.
- A nice article in “The Analysis Factor” describes wide vs long data formats and when to choose which (TLDR, it depends on your statistical analysis plan).
Data cleaning is complicated and varied, and entire books have been written on the subject.910
- For some general considerations on cleaning data, see EDI’s “Cleaning Data and Quality Control” resource
- OpenRefine is an open-source, cross-platform tool for iterative, scripted data cleaning.
- In R, the
tidyverselibraries (e.g.,dplyr,tidyr,stringr) are often used for data cleaning, as are additional libraries likejanitor. - In Python,
pandasandnumpylibraries provide useful data cleaning features. There are also some stand-alone cleaning tools likepyjanitor(started as a re-implementation of the R version) andcleanlab(geared towards machine learning applications). - Both the R and Python data science ecosystems have excellent documentation resources that thoroughly cover data cleaning. For R, consider starting with Hadley Wickham’s R for Data Science book chapter on data tidying,11 and for python check Wes McKinney’s Python for Data Analysis book chapter on data cleaning and preparation.12
Data Analysis
- Harrer, M. et al. Doing Meta-Analysis with R: A Hands-On Guide. 2023. GitHub
- Once again, for R and Python users, the same two books mentioned above provide excellent beginning guidance on data analysis techniques (exploratory analysis, summary stats, visualization, model fitting, etc). In Wickham’s R for Data Science book, the chapter on exploratory data analysis will help. In McKinney’s Python for Data Analysis book, try the chapters on plotting and visualization and the introduction to modeling.
Footnotes
Michener, W.K., Brunt, J.W., Helly, J.J., Kirchner, T.B. and Stafford, S.G. (1997), NONGEOSPATIAL METADATA FOR THE ECOLOGICAL SCIENCES. Ecological Applications, 7: 330-342. https://doi.org/10.1890/1051-0761(1997)007[0330:NMFTES]2.0.CO;2↩︎
Mayernik, M.S. and Acker, A. (2018), Tracing the traces: The critical role of metadata within networked communications. Journal of the Association for Information Science and Technology, 69: 177-180. https://doi.org/10.1002/asi.23927↩︎
Wickham, H. (2014). Tidy Data. Journal of Statistical Software, 59(10), 1–23. https://doi.org/10.18637/jss.v059.i10↩︎
Zimmerman, N. 2016. Hand-crafted relational databases for fun and science↩︎
O’Brien, Margaret, et al. “ecocomDP: a flexible data design pattern for ecological community survey data.” Ecological Informatics 64 (2021): 101374. https://doi.org/10.1016/j.ecoinf.2021.101374↩︎
Wickham, H. (2014). Tidy Data. Journal of Statistical Software, 59(10), 1–23. https://doi.org/10.18637/jss.v059.i10↩︎
Osborne, Jason W. Best practices in data cleaning: A complete guide to everything you need to do before and after collecting your data. Sage publications, 2012.↩︎
Van der Loo, Mark, and Edwin De Jonge. Statistical data cleaning with applications in R. John Wiley & Sons, 2018. https://doi.org/10.1002/9781118897126↩︎
Wickham, Hadley, Mine Çetinkaya-Rundel, and Garrett Grolemund. R for data science. ” O’Reilly Media, Inc.”, 2023. https://r4ds.hadley.nz/↩︎
McKinney, Wes. Python for data analysis. ” O’Reilly Media, Inc.”, 2022. https://wesmckinney.com/book↩︎