Overview
The ltertools package
The goal of ltertools is to centralize the R functions
created by members of the Long Term Ecological Research (LTER)
community. Many of these functions likely have broad relevance that
expands beyond the context of their creation and this package is an
attempt to share those tools and limit the amount of “re-inventing the
wheel” that we each do in our own silos.
The conceptual theme of functions in ltertools is
necessarily broad given the scope of the community we aim to serve. That
said, the identity of this package will likely become more clear as we
accrue contributed functions. This vignette describes the main functions
of ltertools as they currently exist.
Harmonization
The LTER Network is hypothesis-driven with a focus on long term data from sites in the network. This results in data that may reasonably be compared but are–potentially–quite differently formatted based on the logic of the investigators responsible for each dataset. Data harmonization (the process of resolving these formatting inconsistencies to facilitate combination/comparison across projects) is therefore a significant hurdle for many projects using LTER data. We suggest a “column key”-based approach that has the potential to greatly simplify harmonization efforts.
This method requires researchers to develop a 3-column key that contains (1) the name of each raw data file to be harmonized, (2) the name of all of the columns in each of those files, and (3) the “tidy name” that corresponds to each raw column name. Each dataset can then be read in and have its raw names replaced with the tidy ones specified in the key. Once this has been done to all files in the specified folder, they can be combined by their newly consistent column names. A visual version of this column key approach to harmonization is included for convenience here:
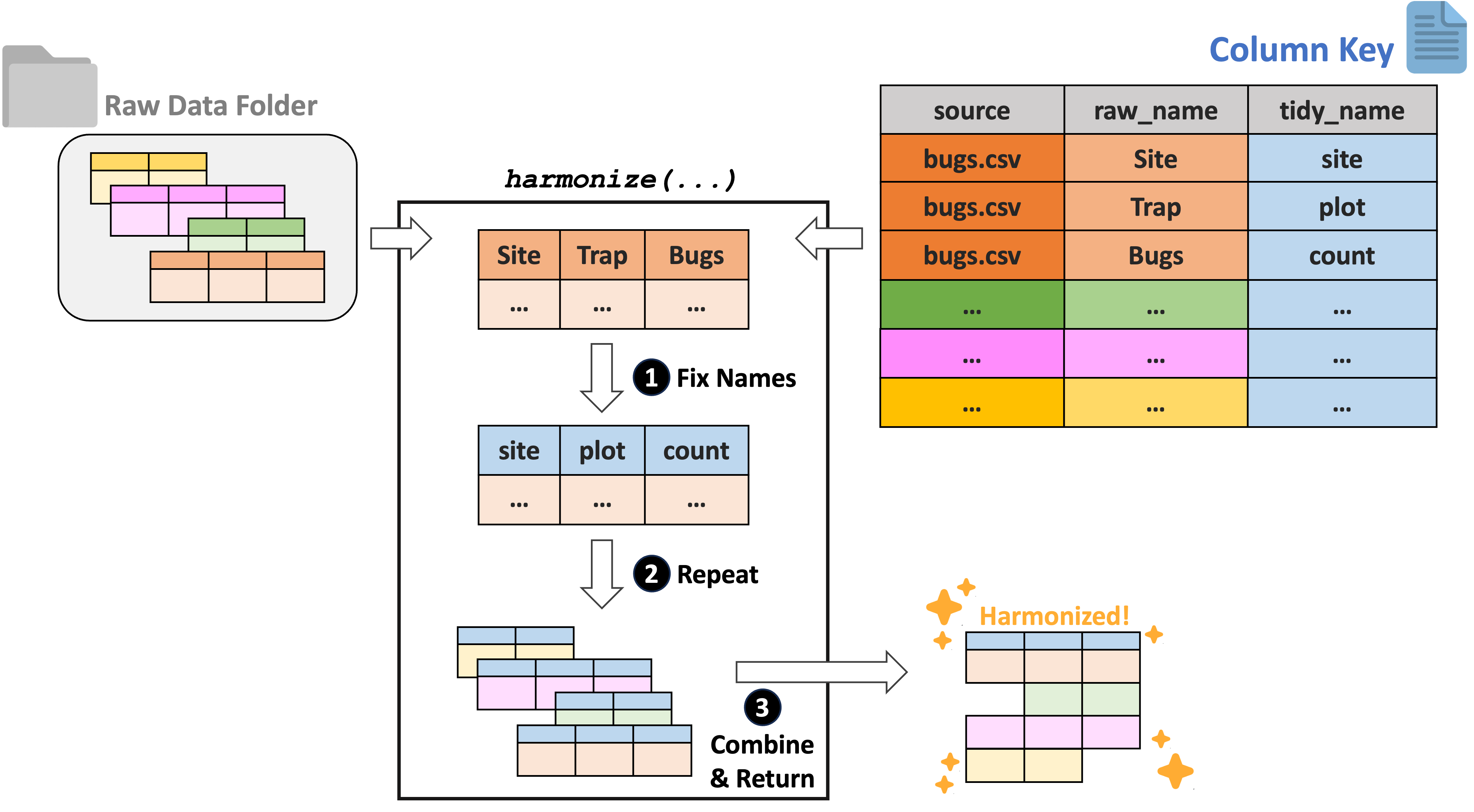
To demonstrate this workflow, we will need to create some example data tables and export them to a temporary directory (so that they can be read back in as is required by the harmonization functions).
# Generate two simple tables
## Dataframe 1
df1 <- data.frame("xx" = c(1:3),
"unwanted" = c("not", "needed", "column"),
"yy" = letters[1:3])
## Dataframe 2
df2 <- data.frame("LETTERS" = letters[4:7],
"NUMBERS" = c(4:7),
"BONUS" = c("plantae", "animalia", "fungi", "protista"))
# Generate a known temporary folder for exporting
temp_folder <- tempdir()
# Export both files to that folder
utils::write.csv(x = df1, file = file.path(temp_folder, "df1.csv"), row.names = FALSE)
utils::write.csv(x = df2, file = file.path(temp_folder, "df2.csv"), row.names = FALSE)While the raw data must be in a folder, the data key is a dataframe in R to allow more flexibility on the user’s end about file format / storage (many LTER working groups like generating their key collaboratively as a Google Sheet). For this example, we can generate a data key manually here.
# Generate a key that matches the data we created above
key_obj <- data.frame("source" = c(rep("df1.csv", 3),
rep("df2.csv", 3)),
"raw_name" = c("xx", "unwanted", "yy",
"LETTERS", "NUMBERS", "BONUS"),
"tidy_name" = c("numbers", NA, "letters",
"letters", "numbers", "kingdom"))
# Check that out
key_obj
#> source raw_name tidy_name
#> 1 df1.csv xx numbers
#> 2 df1.csv unwanted <NA>
#> 3 df1.csv yy letters
#> 4 df2.csv LETTERS letters
#> 5 df2.csv NUMBERS numbers
#> 6 df2.csv BONUS kingdomWith some example files and a key object generated, we can now
demonstrate the actual workflow! The most fundamental of these functions
is harmonize. This function requires the “column key”
described above, as well as the folder containing the raw data
files to which the key refers. Raw data format can also be specified to
any of CSV, TXT, XLS, and/or XLSX. There is a quiet
argument that will silence messages about key-to-data mismatches (either
expected-but-missing column names or unexpected columns).
# Use the key to harmonize our example data
harmony <- ltertools::harmonize(key = key_obj, raw_folder = temp_folder,
data_format = "csv", quiet = TRUE)
# Check the structure of that
utils::str(harmony)
#> 'data.frame': 7 obs. of 4 variables:
#> $ source : chr "df1.csv" "df1.csv" "df1.csv" "df2.csv" ...
#> $ numbers: chr "1" "2" "3" "4" ...
#> $ letters: chr "a" "b" "c" "d" ...
#> $ kingdom: chr NA NA NA "plantae" ...For users that want help generating the column key, we have created
the begin_key function. This function accepts the raw
folder and data format arguments included in harmonize with
an additional (optional) guess_tidy argument. If
TRUE, that argument attempts to “guess” the desired tidy
name for each raw column name; it does this by standardizing casing and
removing special characters. This may be ideal if you anticipate that
many of your raw data files only differ in casing/special characters
rather than being phrased incompatibly. For our example here we’ll allow
the key to “guess”.
# Generate a column key with "guesses" at tidy column names
test_key <- ltertools::begin_key(raw_folder = temp_folder, data_format = "csv",
guess_tidy = TRUE)
# Examine what that generated
test_key
#> source raw_name tidy_name
#> 1 df1.csv xx xx
#> 2 df1.csv unwanted unwanted
#> 3 df1.csv yy yy
#> 4 df2.csv LETTERS letters
#> 5 df2.csv NUMBERS numbers
#> 6 df2.csv BONUS bonusUsers that have already embraced harmonize may want to
add a ‘batch’ of new raw data files to an existing key. In such cases,
begin_key can work but it is cumbersome to need to
manually sort through for only the new rows to add to your existing
column key. The expand_key function exists to simplify this
process. It creates an output much like that of begin_key
but it only includes rows for data files that are not already in (A) the
existing key object or (B) the harmonized data object.
# Make another simple 'raw' file
df3 <- data.frame("xx" = c(10:15),
"letters" = letters[10:15])
# Export this locally to the temp folder too
utils::write.csv(x = df3, file = file.path(temp_folder, "df3.csv"), row.names = FALSE)
# Identify what needs to be added to the existing column key
ltertools::expand_key(key = key_obj, raw_folder = temp_folder, harmonized_df = harmony,
data_format = "csv", guess_tidy = TRUE)
#> source raw_name tidy_name
#> 1 df3.csv xx xx
#> 2 df3.csv letters lettersWrangling
Sometimes it is convenient to read in all of the data files from a
specified folder. read offers the chance to do just that
and returns a list where the name of each element is the corresponding
file name and the contents of the list element is the full data table.
Users may specify the data format or formats they wish to read in.
Currently, read supports CSV, TXT, XLS, and XLSX files.
We can demonstrate this with the test CSVs we created to demonstrate the harmonization workflow earlier.
# Read in all of the CSVs that we created above
data_list <- ltertools::read(raw_folder = temp_folder, data_format = "csv")
# Check the structure of that
utils::str(data_list)
#> List of 3
#> $ df1.csv:'data.frame': 3 obs. of 3 variables:
#> ..$ xx : int [1:3] 1 2 3
#> ..$ unwanted: chr [1:3] "not" "needed" "column"
#> ..$ yy : chr [1:3] "a" "b" "c"
#> $ df2.csv:'data.frame': 4 obs. of 3 variables:
#> ..$ LETTERS: chr [1:4] "d" "e" "f" "g"
#> ..$ NUMBERS: int [1:4] 4 5 6 7
#> ..$ BONUS : chr [1:4] "plantae" "animalia" "fungi" "protista"
#> $ df3.csv:'data.frame': 6 obs. of 2 variables:
#> ..$ xx : int [1:6] 10 11 12 13 14 15
#> ..$ letters: chr [1:6] "j" "k" "l" "m" ...On a different note, the solar_day_info function allows
you to identify the time of sunrise, sunset, and solar noon (as well as
the total length of the day) for each day between a specified start and
end date at a set of latitude/longitude coordinates. All times are in
UTC and the information retrieved is returned as a dataframe.
# Identify day information in Santa Barbara (California) for one week
solar_day_info(lat = 34.41, lon = -119.71,
start_date = "2022-02-07", end_date = "2022-02-12",
quiet = TRUE)
#> date sunrise sunset solar_noon day_length time_zone
#> 1 2022-02-07 2:49:59 PM 1:35:56 AM 8:12:57 PM 10:45:57 UTC
#> 2 2022-02-08 2:49:05 PM 1:36:54 AM 8:13:00 PM 10:47:49 UTC
#> 3 2022-02-09 2:48:10 PM 1:37:52 AM 8:13:01 PM 10:49:42 UTC
#> 4 2022-02-10 2:47:14 PM 1:38:50 AM 8:13:02 PM 10:51:36 UTC
#> 5 2022-02-11 2:46:16 PM 1:39:48 AM 8:13:02 PM 10:53:32 UTC
#> 6 2022-02-12 2:45:17 PM 1:40:45 AM 8:13:01 PM 10:55:28 UTCMuch of the synthesis work with LTER data–and indeed many ecological
research projects generally–requires quantification of
variation. To that end, we’ve written a function that simply
calculates the coefficient of variation (standard deviation divided by
mean) for a vector of numbers. Because sd and
mean both support an argument for defining how missing
values are handled, our cv function does as well.
# Calculate CV (excluding missing values)
ltertools::cv(x = c(4, 5, 6, 4, 5, 5), na_rm = TRUE)
#> [1] 0.1557461We also included a simple function for converting temperature values
(convert_temp) among different accepted units. Simply
specify the values to convert, their current units, and the units to
which you would like to convert and the function will perform the needed
arithmetic. Units are case-insensitive and support either the one-letter
abbreviation or the full name of the unit.
# Convert some temperatures from F to Kelvin
convert_temp(value = c(0, 32, 110), from = "Fahrenheit", to = "k")
#> [1] 255.3722 273.1500 316.4833Note that we chose this function’s naming convention in part to allow for an ecosystem of related ‘unit conversion’ functions that may prove worthwhile to develop.
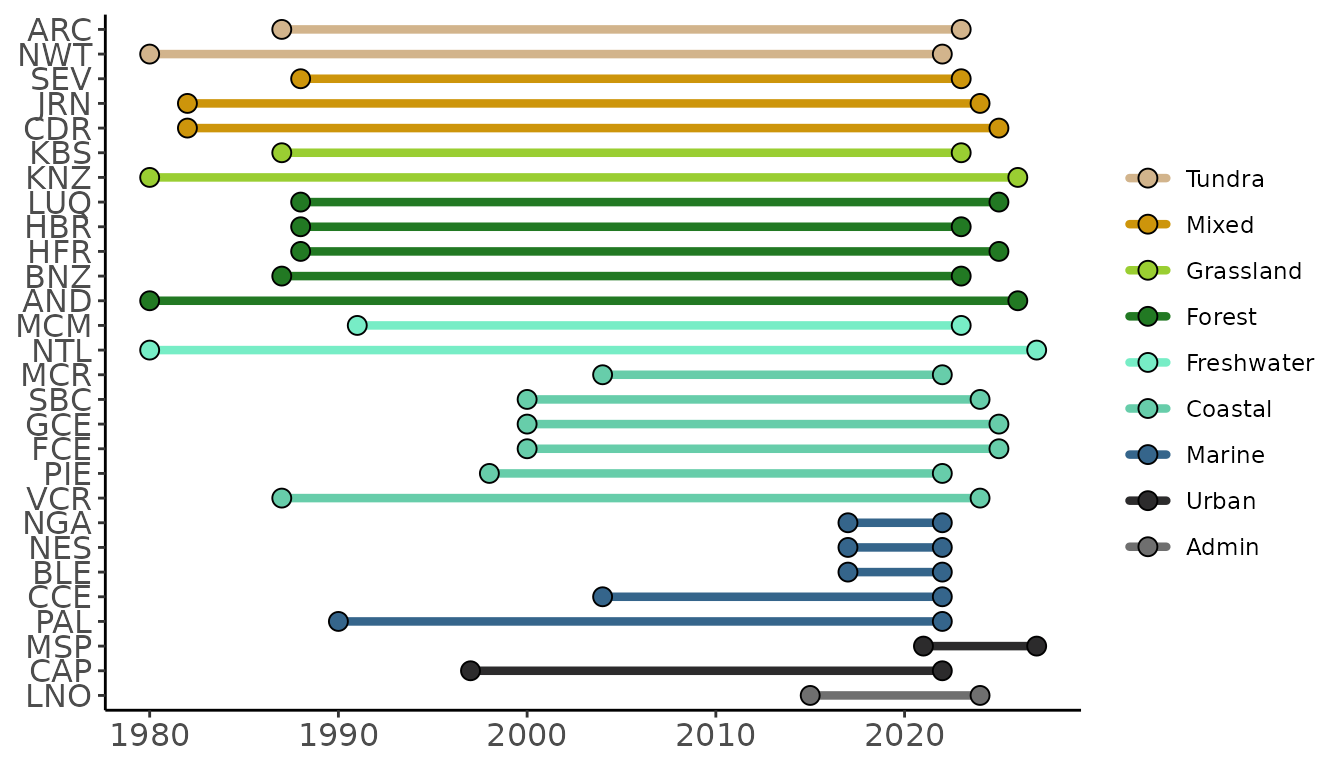
LTER Information
The LTER Network is composed of many separate sites. While all of
these sites are “long term” they do vary slightly in when they were
created. For those interested in knowing the temporal coverage of data
from a particular site or group of sites, site_timeline can
prove a helpful function. This function creates a ggplot2
timeline where sites are on the vertical axis and years are on the
horizontal. Lines are colored based on the habitat of the site and there
is support for a user-defined set of hexadecimal colors though by
default an internal palette is used.
Sites can be specified by their three letter site code or all sites in a particular habitat can be included.
# Check the timeline for all grassland or forest LTER sites
ltertools::site_timeline(habitats = c("grassland", "forest"))
Running the function without specifying site codes or habitat types will result in a timeline of all active LTER sites.
# Check the timeline for all LTER sites
ltertools::site_timeline()